Overview of ImpLiMet
Selecting the optimal imputation method for a dataset depends on evaluating the cause of the missingness in the data and the characteristics of the data. ImpLiMet enables users to impute missing data using 8 different methods and proposes the optimal imputation approach for the user’s data set if users have at least three features and six samples without any missing values in their dataset. ImpLiMet can be used for any dataset. Code is available at: https://github.com/complimet/ImpLiMet
Users can either select an imputation method among the eight provided options (indicated below) or opt for the automated selection of the optimal method for their dataset. The optimized method is determined as follows:
-
ImpLiMet selects the largest subset of data with no missing values from the user’s dataset. There must be at least three features and six samples to optimize the imputation method. If this criteria is not met, the user chooses their own implementation method.
-
Three missing mechanisms are then simulated from the user’s data: missing at random (MAR), missing not at random (MNAR), and missing completely at random (MCAR).
-
Eight imputation methods are used to impute missing data in these three simulations. These imputation methods include five univariate and three multivariate methods:
-
5 univariate methods: mean, minimum, 1/5 minimum, median, and maximum
-
3 multi-variate methods: k-nearest neighbour (KNN), Random Forest (RF), and Multivariate Imputation by Chained Equations (MICE). For the multivariate methods, the required parameters (number of neighbours for KNN, trees for RF, and iterations for MICE) are also optimized.
-
-
The performance of each imputation method is determined by the mean absolute percentage error (MAPE).
-
A comparison table is displayed showing the MAPE for each method. The method with the lowest MAPE across the missingness simulations is suggested as the optimal method. If the full parameter search is selected in the optimization for RF, KNN and MICE, MAPE is calculated for a range of hyperparameters (for RF: 5 to 500 trees, for KNN N from 10 to 100, for MICE 1 to 3 iterations). Without full search selected, only one parameter is tested (for RF, tree of 500, for KNN N=10 and for MICE run uses 2 iterations).
-
The effect of the chosen imputation method on the data structure is assessed and visualized by histograms and principal component analysis (PCA), comparing the impact of removing all features and all samples with missing data to the chosen imputation method on data structure, including kurtosis and skewness information challenging assumptions of a normal distribution.
Input data format and examples as a .CSV file
- Input data without multiple feature measurement groups Column one must contain Sample IDs. Row one must contain feature names.
- Input data with multiple feature measurement groups. If the dataset includes features measured in different units or on different platforms (multiple feature measurement groups), data should be formatted to indicate which feature measurement groups are to be considered separately for missing data simulation. In this case, row one must contain feature names. Row 2 must contain the feature measurement group information.

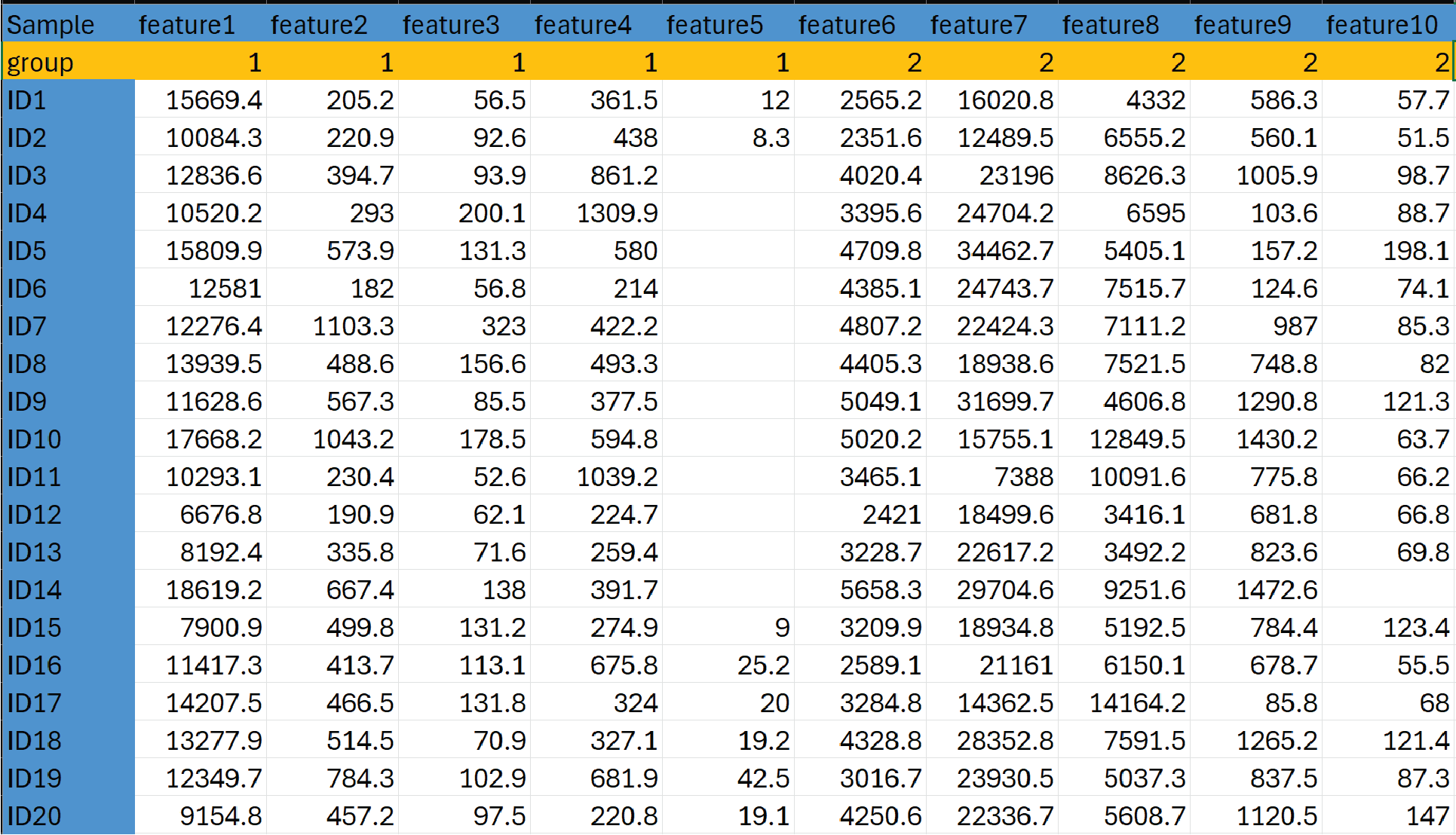
Sample Data
- Input data without multiple feature measurement groups
- Input data with multiple feature measurement groups
Troubleshooting ImpLiMet
When troubleshooting, please review this list of common reasons for ImpLiMet failing to run. If you are still experiencing difficulties, please contact ldomic@uottawa.ca for further assistance. Please include your input dataset and a description of the problem that you experienced. We will reproduce the problem and provide you with a solution.
1. My file does not load or does not produce any results.
ImpLiMet only accepts comma-delimited files as input. Tab-delimited or excel files will be read but will not produce any results. Please convert your input data into .csv format before running ImpLiMet. Data must start from row 2 if there is no feature measurement group information or row 3 if feature measurement group information is selected. Sample information should be listed in the first column. Each feature column must have a unique label.
2. Missing values must be empty strings (cells) or NA.
ImpLiMet's focus is on missing values and recognizes empty strings or NA as missing values. Please convert your missing value indicators to empty strings or NA.
3. ImpLiMet accepts input datasets with any number of feature measurement groups.
Although any number of feature measurement groups is acceptable and will provide results keep in mind that more features in a feature measurement group you have the more accurate will be the imputation.
4. Sample IDs must be unique for each row.
ImpLiMet does not analyze duplicated samples, so sample names must be unique in the input. Please avoid using only numbers for sample naming. Instead, please use a combination of characters and numbers.
5. Selection of an Imputation method provides output but selection of the optimization option crashes.
There must be a minimum of THREE or more complete (no missing values) columns (features) and minimum of SIX complete rows (samples) to run the optimization option. For very small datasets (e.g., dimensions 3x6) the sample and feature threshold must be set to 10% for full optimization.
Contact us
Cite the use of ImpLiMet in a publication
Ou H, Surendra A, McDowell GSV, Hashimoto-Roth E, Xia J, Bennett SAL, Čuperlović-Culf M, (2025) Imputation for Lipidomics and Metabolomics (ImpLiMet): a Web-based application for optimization and method selection for missing data imputation. Bioinformatic Advances , 5, vbae209 doi.org/10.1093/bioadv/vbae209
Public Server
ImpLiMet: https://complimet.ca/shiny/implimet/
Software License
ImpLiMet is free software. You can redistribute it and/or modify it under the terms of the GNU General Public License v3 (or later versions) as published by the Free Software Foundation. As per the GNU General Public License, ImpLiMet is distributed as a bioinformatic tool to assist users WITHOUT ANY WARRANTY and without any implied warranty of MERCHANTABILITY or FITNESS FOR A PARTICULAR PURPOSE. All limitations of warranty are indicated in the GNU General Public License.